- Home
- >
- Core Tech
Uni-MolA Large-Scale Pre-trained Model for 3D Molecular Representation Learning



- Pre-training on billions of 3D molecular structures for representation learning, specifically tailored for property prediction and structural optimization design.
- Encompassing fields such as pharmaceuticals, materials, and chemical engineering, achieving top-class performance in physicochemical, quantitative, and biochemical property predictions.
- Supporting efficient molecular design, empowering practical domains like drug discovery, material optimization, and energy production.
Uni-Mol
Uni-RNAA Large-Scale Pre-trained Model For Nucleic Acids



- We have collected and organized the largest available RNA sequence database to train the most extensive pretraining model for nucleic acids.
- Our model possesses a powerful ability to represent functional structures, consistently outperforming existing algorithms in all known tasks.
- Uni-RNA comprehensively empowers mRNA and nucleic acid drug research and development, accelerating the advancement of new therapeutic approaches.
Uni-RNA
DPA-1A Transferable Interatomic Potential Pre-trained Model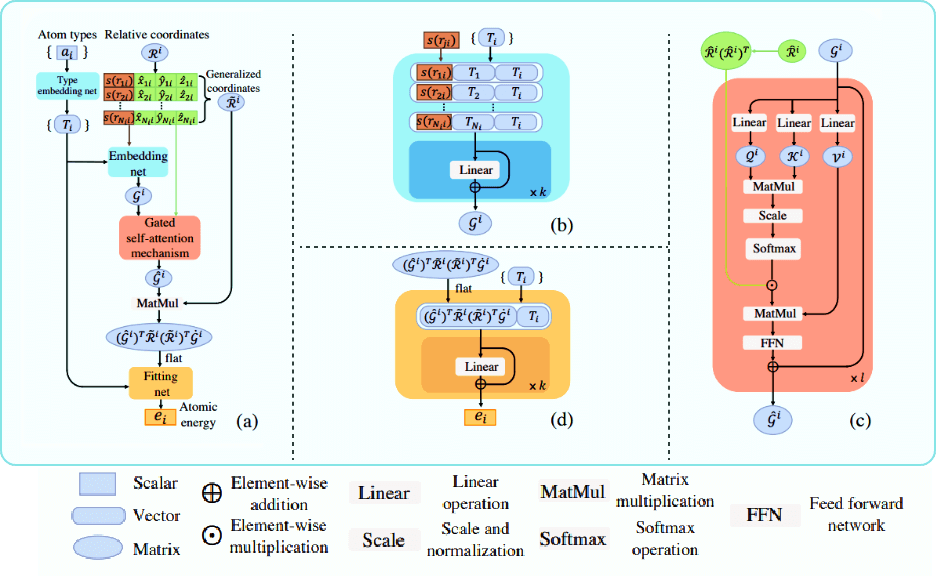



- The world's first pre-trained model covers 70 elements from the periodic table.
- Possesses strong transferability, enabling the fine-tuning of a reliable potential function model with minimal samples on a new system.
- Significantly reduces data dependency for model construction.
DPA-1
Uni-FoldA High-Precision Universal Protein Folding Model



- Unveiling the first open-source Fold model in the domestic market with training code and database, boasting accuracy comparable to AlphaFold2.
- Efficient and precise inference capabilities, supporting complex real-world scenarios such as large proteins, protein complexes, symmetrical proteins, and more.
Uni-Fold
Uni-DockA High-Performance Docking Engine for Large-Scale Database Virtual Screening With GPU
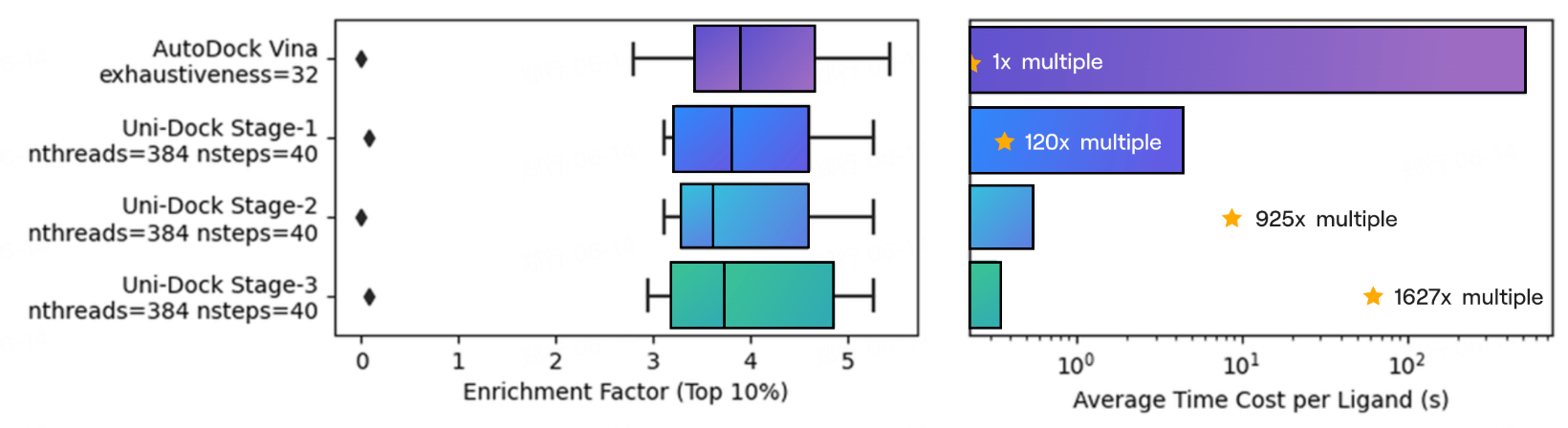


- Maintaining comparable accuracy to traditional molecular docking, Uni-Dock achieves an acceleration rate of over 1600 times on NVIDIA V100 GPU compared to AutoDock Vina's single-core calculation. This performance is more than 10 times faster than other GPU-accelerated molecular docking engines.
- Uni-Dock can complete virtual screening of over 38.2 million molecular databases in less than 12 hours, bringing the virtual screening of large-scale databases, with tens of millions of molecules, into a practical, accessible, and reliable era.
Uni-Dock
